Maintained by Shanliang Zhong
CircPrimer 2.0: a software for annotating circRNAs and predicting translation potential of circRNAs
CircPrimer is a user-friendly tool exploring circRNAs without any special skill required of the users. CircPrimer allows users to search, annotate and visualize circRNAs, and help users to design primers for circRNAs and check specificity of circRNA Primers. CircPrimer can show conserved circRNAs, and predict open reading frames (ORFs), internal ribosomal entry sites (IRESs) and m6A sites. It provides both a graphical and command-line interface.
Download:
| Platform | Java Runtime Environment(JRE) |
| Windows (64-bit) 69.6MB | JRE is not needed. |
| Windows (64-bit) 7.6MB | JRE is needed when running circPrimer2.0. |
| Mac OS X, Linux, and Solaris 3.7MB | JRE is needed when running circPrimer2.0. |
Data Input
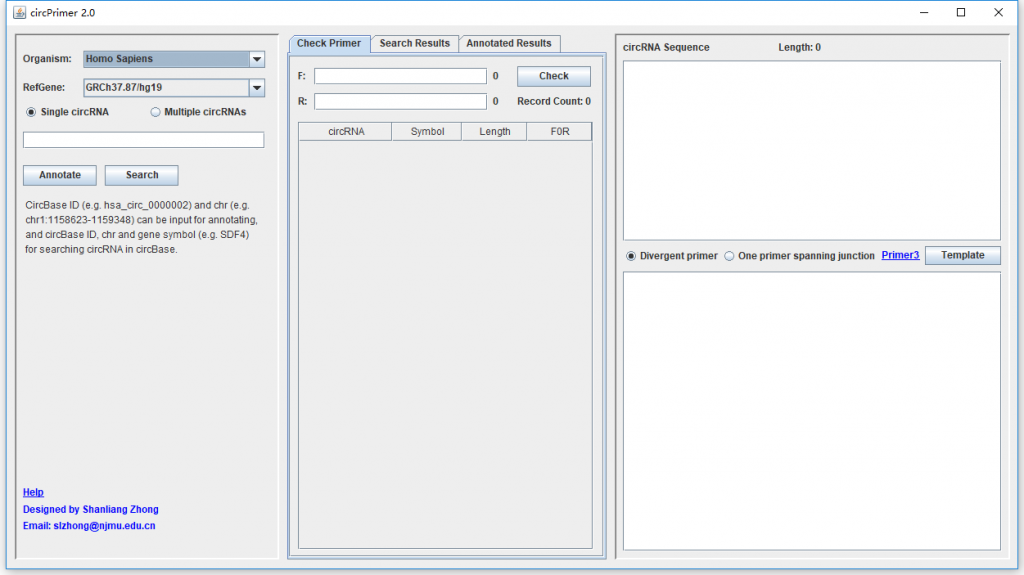
For annotating circRNAs, circBase ID (e.g. hsa_circ_0000002) and genomic location (e.g. chr1:1158623-1159348) are accepted data, and you can check “Single circRNA” or “Multiple circRNAs” to input one or more circRNAs. For searching circRNA in circBase, circBase ID, genomic location and gene symbol (e.g. SDF4) are accepted data.
Searching circRNA in circBase
CircBase ID, genomic location, and gene symbol are the accepted data for searching circRNA in circBase. After inputting the data and clicking “Search” button, the medial panel of “Search Results” will show the results (Figure 1). Click one circRNA in the list, its spliced sequence in circBase will be shown in the topright textarea. If “Annotate circRNA when click” has been checked, a dialog will be present to illustrate the structure of the circRNA (Figure 1). You can save the searching results as different formats by using the right-click menu of the list.
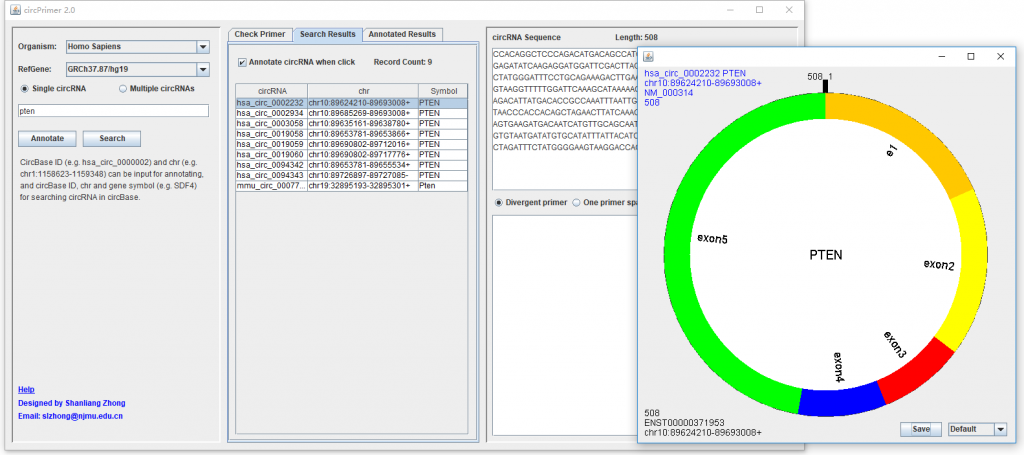
Figure 1. The results of searching “PTEN”.
Annotating circRNAs
CircRNA ID and genomic location are the accepted data for annotating circRNAs. After inputting the data and clicking ‘Annotate’ button, the results will be listed in the medial panel of “Annotated Results”. Click one circRNA in the list, a dialog will be present to illustrate the structure of the circRNA. If “Show RNA” is checked, the annotated spliced sequence will be shown in the topright textarea. If “Show DNA” is checked, the genomic sequence of the circRNA will be shown. You can save the searching results as different formats by using the right-click menu of the list.
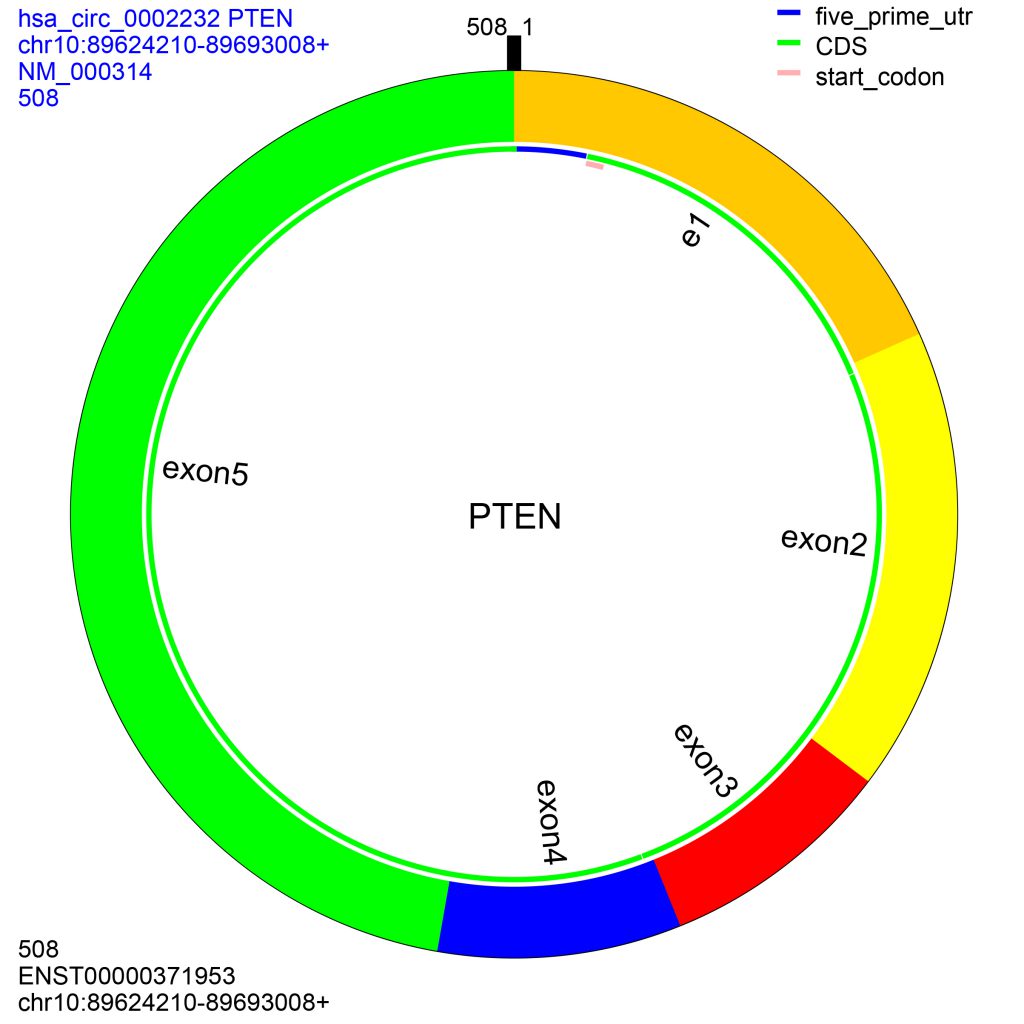
Figure 2. The structure of hsa_circ_0002232. e, partial sequence of the exon; i, partial sequence of the introne. The data on the top-left are from circBase, and the other data are annotated according to their genomic location.
Designing divergent primers
After obtaining spliced sequence as mentioned above, check “Divergent primer” and click “Template” button to generate template for designing divergent primers. Users just paste the generated template to Primer3 to design primers for polymerase chain reaction (PCR).
Designing primers with one primer spanning junction
Use similar method as designing divergent primers, but checking “one primer spanning junction”.
Checking specificity of circRNA primers
To check the specificity of circRNA primers, you can click “Check Primer” panel, input the primers, and click “Check” button to start the checking process. A list will be present to show the circRNAs that can be amplified by the primers. Click one circRNA, a dialog will be present to illustrate the location of the primers (Figure 3). You can save the searching results as different format by using the right-click menu of the list.
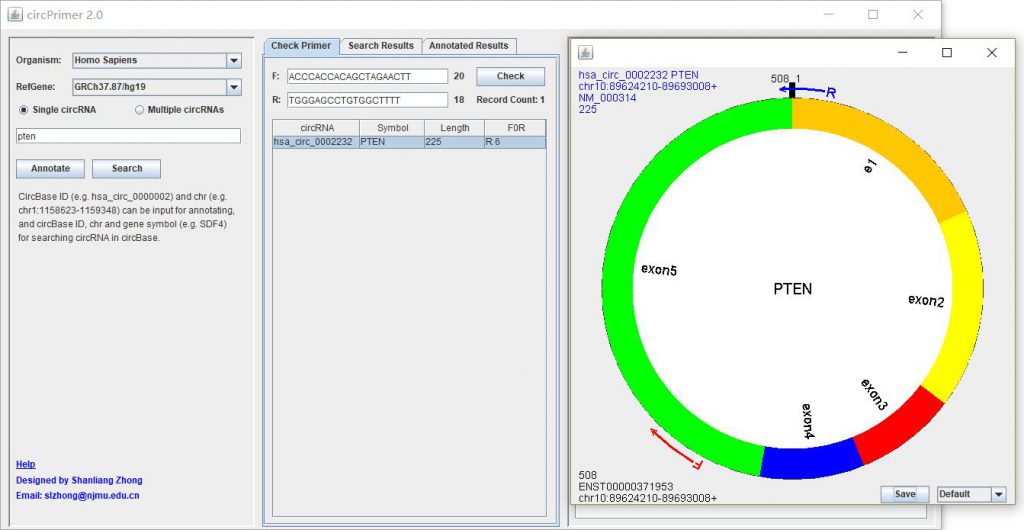
Figure 3. Checking specificity of circRNA primers and showing the location of the primers. R0F indicates primer type: 0, divergent primer; F/R + number, “F” indicates forward primer spanning the junction, “R” indicates reverse primer, and “number” indicates the base number of the primer 3` end spanning the spliced junction.
Predicting open reading frames (ORFs)
When the dialog is present to show the circRNA structure, you can select bottom-right ComboBox as “ORFs” to show the predicted ORFs. You can select one or more ORFs to show detailed information. ORFs may be in the same frame as their parental genes, thus they could code same amino acid fragments. circPrimer2.0 marks these ORF with red font, and the amino acid fragments are highlighted with underline (Figure 4).
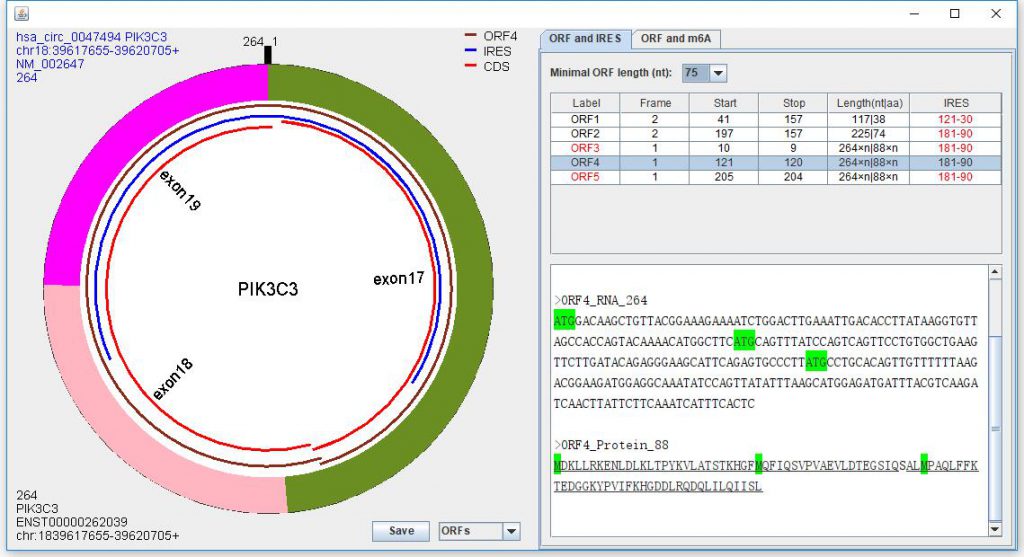
Figure 4. Predicting open reading frames (ORFs) and internal ribosomal entry site (IRES). ORF3~ORF5 are infinite ORFs which lack stop codon and labeled with “a number×n” in the Length field. The number is the length of one repeat sequence. Red font in IRES field is used to highlight the IRESs spanning the junctions. Red font in ORF field is used to highlight the ORFs coding same amino acid fragments as their parental genes; the amino acid fragments are highlighted with underline; and CDS represents the ORF region in parental gene.
Predicting internal ribosomal entry site (IRES)
When showing the predicted ORFs, the predicted result of IRESs also be shown. If IRES field is labeled as “None”, it means that none IRES is found in this circRNA. If two or more IRESs are predicted, the IRES near the ORF start is considered as the IRES of this ORF (Figure 4).
Predicting ORFs and IRES with a circRNA sequence
Input sequence to the top-right textarea and use the right-click menu to predict ORFs and IRESs (Figure 5).
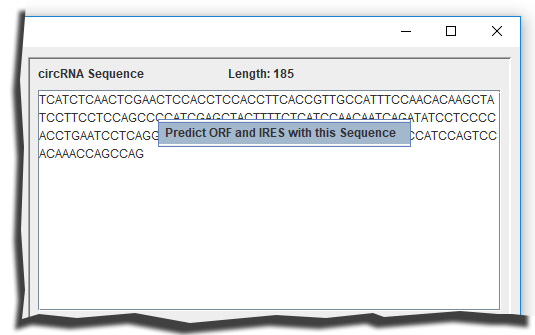
Figure 5. Predicting ORFs and IRESs with a circRNA sequence.
Showing m6A modification sites on circRNA
After showing the predicted ORFs, you can click “ORF and m6A” to show the potential m6A sits on this circRNA (Figure 6). The “High-confidence” are the modifications from m6A-Atlas database. “DRACH” and “RRACH” are potential m6A modification sites matching “DRACH” and “RRACH” (D=A, G or U; R = G or A; H = A, C or U).
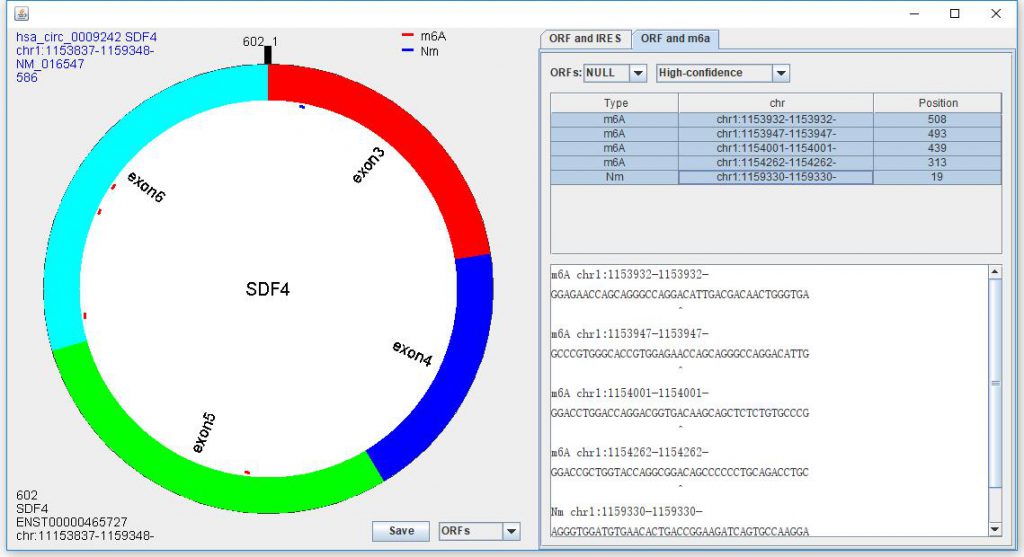
Figure 6. Showing ORF and m6A modification sites.
Showing homeotic gene
In the middle panel, the listed circRNAs in red font are conserved circRNAs. When clicking these circRNAs, the bottom-right textarea will show the identity between Homo sapiens and Mus musculus.
Command-line interface
(1) Predicting ORFs
java -jar circPrimer2.0.jar ORF min_ORF_len input_path output_path
(2) Predicting IRESs
java -jar circPrimer2.0.jar IRES step IRESlength input_path output_path
(3) Predicting ORFs and IRESs
java -jar circPrimer2.0.jar OI min_ORF_len step IRESlength input_path output_path
ORF, predicting ORFs only; min_ORF_len, minimal ORF length; input_path, path of a FASTA file; output_path, output file name; IRES, prodicting IRESs only; step, step used to split the sequence; IRESlength, length of subsequences used to predict IRES; OI, predicting both ORFs and IRESs.
How to cite?
1. Shanliang Zhong, Jifeng Feng, CircPrimer 2.0: a software for annotating circRNAs and predicting translation potential of circRNAs, BMC Bioinformatics, 2022, 23(1): 215
2. Shanliang Zhong, Jinyan Wang, Qian Zhang, Hanzi Xu, Jifeng Feng, CircPrimer: a software for annotating circRNAs and determining the specificity of circRNA primers, BMC Bioinformatics, 2018, 19(1): 292
Download Former Version: